Imaging algorithm visualizes early heart abnormalities
Studying abnormalities that occur during early cardiac development is very important for understanding congenital heart defects. Optical coherence tomography (OCT) is a noninvasive imaging modality based on low-coherence interferometry that is capable of micrometer-scale resolution and 1–2mm of depth penetration in embryonic tissue. OCT can visualize and quantify real-time heart dynamics in 3D, making it a useful tool for assessing cardiac development. Real-time volume imaging performed with current systems, however, does not provide the necessary temporal resolution to analyze rapid cardiac dynamics in 3D.
In the past, researchers used a physiological signal such as an electrocardiogram to trigger data acquisition, typically during multiple phases of the cardiac cycle.1–4 Alternatively, some recorded the physiological signal while imaging as fast as possible over several heartbeats at different spatial locations, and then used timing from the signal to create 3D volumes over time.1,5–8 However, recording an additional signal is invasive and time-consuming. A possible remedy is retrospective cardiac gating,9–12 where one obtains data over multiple heartbeats without recording an external signal and rearranges the data after the fact to reconstruct 3D volumes over time. Our technique, called image-based retrospective gating, employs image similarity to rearrange out-of-order image data. The result is extremely high temporal resolution.
Our image-based retrospective gating algorithm is influenced by the work of Liebling et al.10 However, due to the higher signal-to-noise ratio of our imaging setup, we work in the spatial rather than the wavelet transform domain. In addition, we created a novel quantitative validation technique to obtain a real-time reference scan of the same heart volume used for out-of-order data acquisition. We then employ it to evaluate the accuracy of our algorithm.
First we asynchronously image a few heartbeats worth of data at equally spaced steps (slice positions) along the heart volume. To retrospectively gate the acquired data we perform spatial data decimation, cardiac cycle period estimation using an optimization approach with image similarity measures, and data reassembly based on cardiac cycle period estimates. We also perform linear interpolation in time of reassembled data, relative shift estimation between time series of 2D image frames from adjacent slices in volume, consolidation of relative shifts to produce absolute shift estimates, and data reordering to create 3D volumes over time. We also created a novel quantitative validation technique to obtain a real-time reference scan of the same heart and perform cross-correlation image matching between frames from the reference scan and the gated data. We then plot matching volume numbers in gated data against slice position. We assess the linearity of the plot to evaluate the accuracy of retrospective gating.
We applied our algorithm to out-of-order data collected from a stage 14 embryonic quail heart (120 frames, 4.27ms frame interval, ~234 frames/s) at 80 slice positions along the heart volume with our Fourier domain mode-locked OCT system. Using retrospective gating, we obtained 90 3D volumes across a single cardiac cycle, making it possible to visualize 3D systolic dynamics of the beating avian embryo heart for the first time. Figure 1 shows single frames from three movies of the beating heart with cutaway views in sagittal (en face), coronal (OCT imaging plane), and transverse (along the heart tube) planes. For the first time, we saw visually perceptible differences in contraction patterns at the inflow and outflow of the embryonic heart tube, which was impossible with lower temporal resolution scans: see Figure 1(c).

We also demonstrated our quantitative validation experiment using two real-time, reference full-volume scans. Figure 2 shows the plot of matching volume numbers (obtained by cross-correlation image matching) from the gated data against spatial slice position for scans A and B. We easily obtained a linear fit through these data points. The figure shows that the slopes of these linear fits closely match the ideal slope of the reference scan (1.275ms/slice), demonstrating the accuracy of our gated reconstructions.
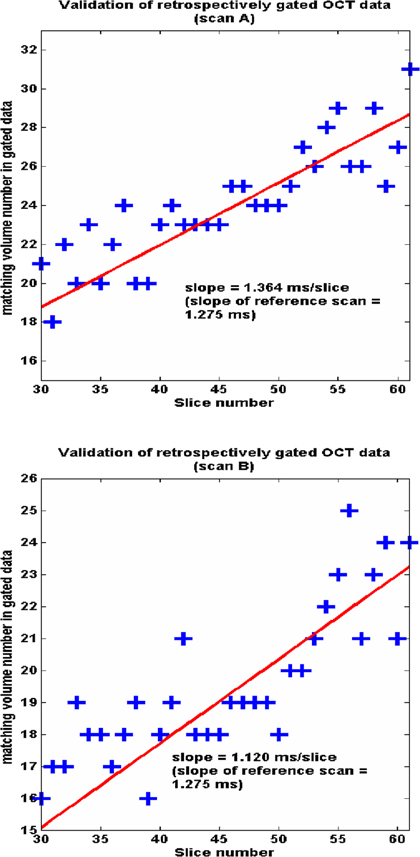
To conclude, we developed an algorithm for image-based retrospective gating of OCT image data and applied it successfully on out-of-order (2D plus time) data collected from the early embryonic quail heart to obtain a series of heart volumes (3D plus time). We proposed a robust validation scheme for quantitative evaluation of our proposed algorithm, which has revealed that errors in reconstruction (4.7ms standard deviation) are small enough to visualize 4D cardiac systolic dynamics for the first time. The extremely high effective temporal resolution enabled by our gating algorithm will allow us to track blood cells, measure minute and rapid tissue displacements, and make biomechanical measurements such as shear stress on the heart wall. This in turn will expand our understanding of the mechanical forces that regulate tissue function in the looping embryonic heart.
Going forward, we will use this new technology to study how the structure and function of the heart are tied together to control development at early stages. Future improvements will focus on precisely measuring stresses on tissue and studying the mechanotransduction signaling that influences heart development.
This research is supported by funds from the National Institutes of Health (1RO1HL083048).
